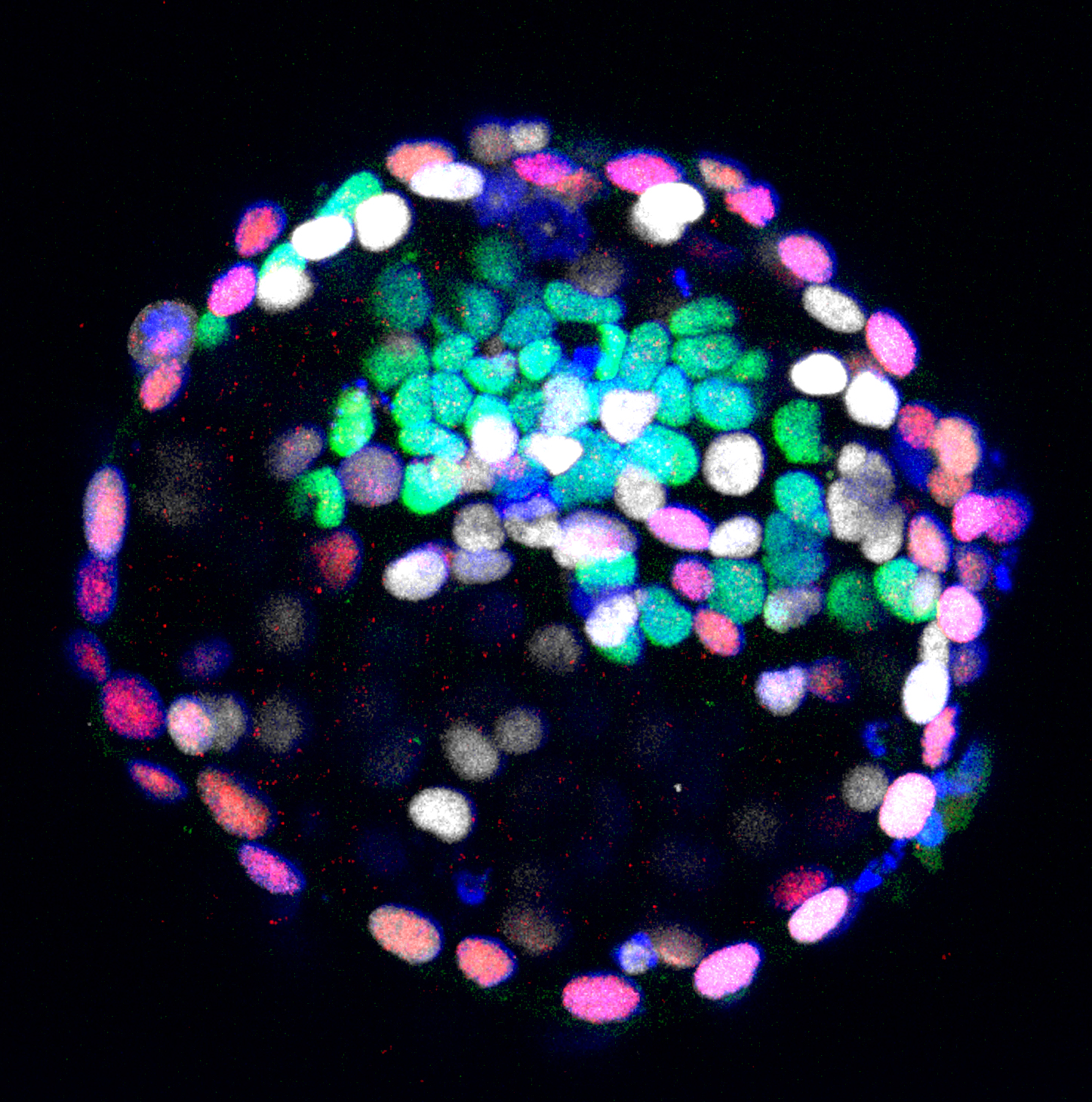
Welcome to the Laboratory for Primate Embryogenesis!
Our lab focuses on how embryonic cells organise themselves to form the most complex lifeforms, such as human and non-human primates.
We follow primate embryonic cells through parts of their journey to provide insights into human development. Our approaches include simultaneous genetic and epigenetic high-throughput sequencing from single cells, embryonic stem cell culture and bioengineering of stem cell-based embryo models.
A deeper understanding of primate development is vital for innovative treatments of implantation failure, infertility and cancer as well as clinical applications of stem cell biology.
Biography
Thorsten originates from Austria, where he studied Molecular Biology at the University of Vienna. After completion of his master’s thesis in 2007, he was awarded a PhD-fellowship at the University of Sheffield to work on neuronal differentiation of mouse embryonic stem cells. Thorsten early postdoctoral work in the laboratories of Prof. Austin Smith and Prof. Jennifer Nichols provided transcriptional and functional evidence that the closest in vivo counterpart of mouse embryonic stem cells is the preimplantation epiblast. Between 2012 and 2015, he pioneered genome-wide comparison of mouse to non-human primate development by lineage-specific RNA-seq, which identified a primate-specific role for WNT signaling during early lineage specification. In 2017, Thorsten was awarded a Sir Henry Dale Fellowship to start his own group – the Laboratory for Primate Embryogenesis – at the Department of Physiology, Development and Neuroscience at the University of Cambridge. Thorsten is a member of the Cambridge Stem Cell Institute, the Centre for Trophoblast Research, the Anne McLaren Trust Fund and Fellow of Darwin College.
Research
Embryo implantation, gastrulation and organogenesis
The first signs of the human body axis can be traced back to the second week of gestation. To get to this point, the blastocyst implants and establishes a small sheet of cells, the embryonic disc. Deeply embedded within extraembryonic tissues, the embryo undergoes a reorganization process termed “gastrulation”, which transforms the embryonic disc into three germ layers and determines the entire future body plan.
Most of our knowledge on mammalian gastrulation is based on mouse, but human embryogenesis differs in anatomical architecture, timing, molecular configuration and sequence of cell-fate decisions. For instance, in human and non-human primates, the implanting epiblast polarises into a rosette, gives rise to amnion and forms a flat EmDisc. Rodent embryos form amnion later in development, after gastrulation.
Research goals
The central aim of our research focuses on delineating the molecular crosstalk between the human embryonic disc and extraembryonic signalling centres, which control the sequence of cell-fate allocation in the primitive streak. To understand how these signalling centres emerge, we need to elucidate how extraembryonic lineages are specified in primate postimplantation development.
Our lab was the first to reveal the molecular landscape in primate embryos between implantation and gastrulation in vivo (Bergmann et al., Nature 2022). We developed epiblast- and amnion-spheroid cultures using microfluidics (Schindler et al., Stem Cell Reports 2021; Munger et al., Development 2022) and pioneered computational approaches, including spatial-identity-mapping, to determine the identity of in vitro cultured cells. Our ongoing work involves micropatterning, blastoids, microfluidics and bioprinting to emulate human and non-human primate development in stem cell-based embryo models.
The results from our work will be critical to understand human implantation failure, how errors in gastrulation can lead to congenital malformations and how germ layers are patterned for organ formation.
Spatial transcriptome profiling of primate embryos - view the video
Collaborators
Prof. Erika Sasaki (Central Institute for Experimental Animals, Tokyo, Japan)
Prof. Ruediger Behr (German Primate Centre, Germany)
Prof. Jan Brosens (University of Warwick, UK)
Prof. Florian Hollfelder (Department of Biochemistry, Cambridge, UK)
Prof. Kathy Niakan (Centre for Trophoblast Research, Cambridge, UK)
Dr. Peter Rugg Gunn (Babraham Institute, Cambridge, UK)
Prof. Wolf Reik (Altos Labs, Cambridge, UK)
Publications
Castillo-Venzor A, Penfold CA, Morgan MD, Tang WW, Kobayashi T, Wong FC, Bergmann S, Slatery E, Boroviak TE, Marioni JC, Surani MA. Origin and segregation of the human germline. Life Sci Alliance, 2023
Kraunsoe S, Azami T, Pei Y, Martello G, Jones K, Boroviak TE, Nichols J. Requirement for STAT3 and its target, TFCP2L1, in self-renewal of naïve pluripotent stem cells in vivo and in vitro. Biology Open, 2023.
Sheng G, Boroviak TE, Schmidt-Ott U, Srinivas S. Extraembryonic tissues: exploring concepts, definitions and functions across the animal kingdom. Philosophical Transactions of the Royal Society B, 2022
Siriwardena D, Boroviak TE. Evolutionary divergence of embryo implantation in primates. Philosophical Transactions of the Royal Society B, 2022
Munger C, Kohler TN, Slatery E, Ellermann AL, Bergmann S, Penfold CA, Ampartzidis I, Chen Y, Hollfelder F, Boroviak TE. Microgel culture and spatial identity mapping elucidate the signaling requirements for primate epiblast and amnion formation. Development, 2022
Chen Y, Siriwardena D, Penfold C, Pavlinek A, Boroviak TE. An integrated atlas of human placental development delineates essential regulators of trophoblast stem cells. Development, 2022
Malkowska A, Penfold C, Bergmann S, Boroviak TE. A hexa-species transcriptome atlas of mammalian embryogenesis delineates metabolic regulation across three different implantation modes. Nature Communications, 2022
Bergmann S, Penfold CA, Slatery E, Siriwardena D, Drummer C, Clark S, Strawbridge SE, Kishimoto K, Vickers A, Tewary M, Kohler TN, Hollfelder F, Reik W, Sasaki E, Behr R, Boroviak TE. Spatial profiling of early primate gastrulation in utero. Nature, 2022.
Boroviak TE. A human embryo model cracks symmetry breaking. Cell Stem Cell, 2022
Bergmann S, Schindler M, Munger C, Penfold CA, Boroviak TE. Building a stem cell based primate uterus. Nature Communications Biology, 2021.
Schindler M, Siriwardena D, Kohler TN, Ellermann AL, Slatery E, Munger C, Hollfelder F, Boroviak TE. Agarose microgel culture delineates lumenogenesis in naive and primed human pluriopotent stem cells. Stem Cell Reports, 2021
Betto RM, Diamante L, Perrera V, Audano M, Rapelli S, Lauria A, Incarnato D, Arboit M, Pedretti S, Rigoni G, Guerineau V, Touboul D, Stirparo GG, Lohoff T, Boroviak T, Grumati P, Soriano ME, Nichols J, Mitro N, Oliviero S, Martello G. Metabolic control of DNA methylationin naive pluripotent cells. Nature Genetics, 2021.
Stirparo GG, Kurowski A, Yanagida A, Bates LE, Strawbridge SE, Hladkou S, Stuart HT, Boroviak TE, Silva JCR, Nichols J. OCT4 induces embryonic pluripotency via STAT3 signalling and metabolic mechanisms. PNAS, 2021
Connor R. and Boroviak T, Origin and function of the yolk sac in primate embryogenesis, Nature Communications, 2020
Boroviak T, Stirparo GG, Dietmann S, Hernando-Herraez I, Mohammed H, Reik W, Smith A, Sasaki E, Nichols J, Bertone P, (2018), Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development, Development 2018
Stirparo GG, Boroviak T, Guo G, Nichols J, Smith A, Bertone P (2018), Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast, Development 2018
Boroviak T and Nichols J, (2017), Postimplantation development predicts extraembryonic potential of naive pluripotency in primates, Development, 2017
Scognamiglio R, Cabezas-Wallscheid N, Their M, Altamura S, Reyes A, Baumgärtner D, Prendergast A, Carnevalli L, Paleske L, Boroviak T, Wörsdörfer P, Essers M, Eisenman R, Edenhofer F, Bertone P, Huber W, Hoeven F, Smith A and Trumpp A, Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause, Cell, 2016
Boroviak T*, Loos R*, Lombard P, Behr R, Sasaki E, Nichols J, Smith A and Bertone P, (2015), Lineage-specific profiling delineates the emergence and progression of naïve pluripotency in mammalian embryogenesis, Developmental Cell, 2015
Boroviak T. and Nichols J, (2015), Maximising clonal embryonic stem cell derivation by ERK pathway inhibition, Methods in Molecular Biology, 2015
Boroviak T. and Nichols J, (2014), The birth of embryonic pluripotency, Philosophical transactions of the Royal Society 2014
Boroviak T, Loos R, Bertone P, Smith A and Nichols J, The ability of inner cell mass cells to self-renew as embryonic stem cells is acquired upon epiblast specification, Nature Cell Biology 2014
Tatsumoto S, Adati N, Tohtoki Y, Sakaki Y, Boroviak T, Habu S, Okano H, Suemizu H, Sasaki E and Satake M, (2013), Development and characterization of cDNA resources for the common marmoset: one of the experimental primate models, DNA Res. 2013
Boroviak T. and Rashbass P, (2010), The Apical Polarity Determinant Crumbs 2 is a Novel Regulator of Embryonic Stem Cell Derived Neural Progenitors, Stem Cells 2010
Reitinger S, Boroviak T, Laschober GT, Fehrer C, Muellegger J, Lindner H, Lepperdinger G, (2008), High-yield recombinant expression of the extremophile enzyme, bee hyaluronidase in Pichia pastoris, Protein Expr Purif. 2008